About SMARTCyp
What does SMARTCyp do?
SMARTCyp is a method for prediction of which sites in a molecule that are most liable to metabolism by Cytochrome P450. It has been shown to be applicable to metabolism by the isoforms 1A2, 2A6, 2B6, 2C8, 2C19, 2E1, and 3A4 (CYP3A4), and specific models for the isoform 2C9 (CYP2C9) and isoform 2D6 (CYP2D6) are included from version 2.1. CYP3A4, CYP2D6, and CYP2C9 are the three of the most important enzymes in drug metabolism since they are involved in the metabolism of more than half of the drugs used today.
The methodology
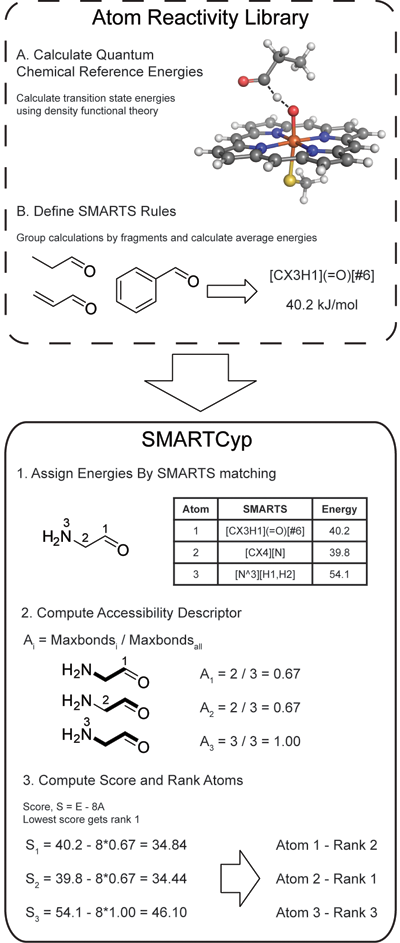
The SMARTCyp method is described in the figure to the right. To construct the program a large number of quantum chemical calculations of fragment activation energies have been performed. The results have been compiled into rules consisting of SMARTS patterns and associated energies.
SMARTCyp only uses the 2D structure of a compound, and the energy required for oxidation at each atom is computed by fragment matching towards the SMARTS patterns. If multiple patterns match the same atom the pattern with the lowest energy is used. The accessibility is approximated as the relative topological distance of an atom from the center of the molecule, and the final score is computed as Score = energy - 8*accessibility.
The SMARTCyp 2D6 model is slightly more complex, using the basic SMARTCyp reactivity, plus the distance from an atom to the end of the molecule (Span2End), and the distance from an atom to a protonated amine nitrogen atom (N+dist). A linear correction is then applied giving a final 2D6-score = energy + 6.7*(8 - N+dist + Span2End). Cutoffs are applied so that N+dist is never larger than 8, and Span2End is never larger than 4.
The SMARTCyp 2C model is a remake of the 2D6 model, using the basic SMARTCyp reactivity, plus the distance from an atom to the end of the molecule (Span2End), and the distance from an atom to a carboxylic acid (or a bioisoster thereof) oxygen atom (COO-dist). A linear correction is then applied giving a final 2C-score = energy + 5.9*(8 - COO-dist + Span2End). Cutoffs are applied so that COO-dist is never larger than 8, and Span2End is never larger than 4.
There is also a correction for unlikely N-oxidations (from version 2.3). A set of tertiary alkylamines for which the reactivity is a bad representation of the likelyhood for the generation of an N-oxide are given a penalty of 100 kJ/mol to their nitrogen atom's energy. A detailed description of why this is scientifically sound can be seen in the paper where we find that they are actually limited by their N-inversion barrier.
The implementation has been done as a java program using the CDK and JChemPaint java libraries. Input files of formats other than sdf, mol and smi (which can be handled directly by the java program) are converted to sdf format using OpenBabel.
The method has been published as SMARTCyp: a 2D-method for Prediction of Cytochrome P450 Mediated Drug Metabolism in ACS Medicinal Chemistry Letters.
SMARTCyp 2.4 performance
Since the SMARTCyp method is built from computational data which is not isoform dependent it is in principle applicable to all CYP isoforms. However, since the properties of the active site can affect the binding conformation of substrates there are most likely isoforms for which the simple SMARTCyp method breaks down. The standard model in SMARTCyp has been validated against CYP3A4 substrates since CYP3A4 is the isoform which is most promiscuous with regard to substrate shape and size.
We have recomputed the accuracies of SMARTCyp 2.4 on the data sets from RS-Predictor models augmented with SMARTCyp reactivities: Robust metabolic regioselectivity predictions for nine CYP isozymes, and the results are shown below. For all isoforms except 2C8, 2C9, 2C19 and 2D6 the standard SMARTCyp model has been used.
| 1A2 | 2A6 | 2B6 | 2C8 | 2C9 | 2C19 | 2D6 | 2E1 | 3A4 | |
|---|---|---|---|---|---|---|---|---|---|
| Rank 1 (%) | 65 | 72 | 66 | 68 | 71 | 70 | 74 | 64 | 65 |
| Rank 1-2 (%) | 80 | 86 | 77 | 83 | 84 | 86 | 83 | 82 | 78 |
| Rank 1-3 (%) | 88 | 91 | 87 | 91 | 91 | 91 | 90 | 89 | 85 |
Understanding the table above: For example, the number 81 for rank 1-2 in the Any metabolite column means that for 81% of the compounds tested, an experimental metabolic position was found among the top 2 atoms predicted by SMARTCyp.
Job execution and storage info
The program is exectuted directly upon job submission, and usually takes only a few seconds to run. For files with many molecules the time required is the number of molecules divided by three seconds (as long as only one job is running on the server at this time).
At the end of each month the output of all jobs are deleted.
Development
The science and development of SMARTCyp is performed at the Department of Medicinal Chemistry at the University of Copenhagen, in the group of Biostructural Research. The people working on the program are Patrik Rydberg (QC calculations, model building, java programming, web service), David Gloriam (java programming) and Lars Olsen (project leader).
Funding
The current development of the SMARTCyp program and the research behind the models is funded by Lhasa Limitied.
The initial development and first versions of SMARTCyp (until version 1.5.3) was funded by the Alfred Benzon foundation and the Danish Council for Independent Research.